In Vitro Evaluation of Hepg2 Cell Proliferation Altered by Reactive Oxygen and Nitrogen Species
Pengcheng Li
DOI10.21767/2572-309X.1000112
Pengcheng Li*
The Medical School, The University of Queensland, QLD, Australia
- Corresponding Author:
- Pengcheng Li
The Medical School, The University of Queensland, QLD 4006, Australia.
Tel: 0061416183106
E-mail: pengcheng.li@uqconnect.edu.au
Received date: April 16, 2016; Accepted date: May 17, 2016; Published date: May 24, 2016
Citation: Li P. In Vitro Evaluation of Hepg2 Cell Proliferation Altered by Reactive Oxygen and Nitrogen Species. J. Adenocarcinoma. 2016, 1:2. doi: 10.21767/2572-309X.100012
Abstract
This study was to investigate cell proliferation regulated by reactive oxygen and nitrogen species and their scavengers. Earlier conclusions are paradoxical on roles that reactive oxygen and nitrogen species and their scavengers enhance or inhibit cancer cell proliferations. This study employed the MTS assay to evaluate proliferations of HepG2 cells treated with various concentrations of two different nitric oxide donors, hydrogen peroxide and their scavengers for 24 hours. The MTS assay revealed that low concentrations of SNP, SNAP and H2O2 can significantly enhance HepG2 cell proliferation, and high concentrations of cPTIO significantly inhibited HepG2 cell proliferation. Additionally, the assay also indicated that 100 μM H2O 2 , 100 μM cPTIO or 100 U/mL catalase did not have evident synergisms with NSP or SNAP. However, due to reactions of the chemicals with reagents of kits of the MTS assay, the data were not inferred that low concentrations of NO and H2O2 enhance proliferation, whereas high concentrations inhibit proliferation, and whether there are synergistic effects with the combination of SNP (or SNAP) and H2O2 , cPTIO or catalase.
Keywords
Hepatocellular carcinoma; Reactive oxygen species; Reactive nitrogen species; Cell proliferation; Synergism
Introduction
The hallmark of hepatocellular carcinoma (HCC) is uncontrolled proliferation of tumour cells, which ignore inhibitory signals to cell proliferation, promote growth of blood vessels and invade other organs [1]. The detailed mechanisms for this are still unclear [2,3]. Mounting evidence suggests that reactive oxygen species (ROS) and reactive nitrogen species (RNS) generated through chronic liverinflammation are involved in the process of HCC development [4-6]. Therefore, a precise understanding how ROS and RNS regulate cell proliferation will help decipher the enigma of hepatocarcinogenesis.
Epidemiology has revealed that most of HCCs develop in chronic inflammatory conditions of the liver [1]. In these conditions, various activated inflammatory and immune cells are recruited to the site of inflammation and generate ROS and RNS. Local concentrations of ROS and RNS have been shown to be elevated in chronic liver inflammation [7-10].
As described in literature reviews, ROS and RNS can interact with nucleic acids, proteins and other biological macromolecules to abnormally regulate cell proliferation. For example, ROS and RNS are able to oxidize DNA to induced genetic instability [11-13]. This may randomly induce mutations of key genes in hepatocytes like tumour suppressor genes or oncogenes resulting in a growth advantage for affected cells that ultimately leads to abnormal regulation of cell proliferation and development of HCCs [14,15]. ROS and RNS may also regulate cell proliferation through changes in key amino acids in proteins such as oxidation [16], S-nitrosylation of thiols of cysteines [17-19] or nitration of tyrosine [20,21].
Cell proliferation is perhaps influenced by concentrations of ROS and RNS, with the potential for synergisms effects between NO and ROS. Reports revealed that low concentrations of ROS and RNS can enhance cell proliferation, whereas high concentrations have been shown to inhibit cell growth [5,22-24]. For example, this dual role of H2O2 has also been observed in previous studies with keratinocytes [25], HeLa cells [26], lens epithelial cells [27], human prostate cancer cells [28] and cardiomyocytes [29]; with cell viabilities assessed using the MTS or MTT assay. Likewise, NO also exerts the dual role that low concentrations of NO enhance proliferations of head and neck squamous cell carcinoma FaDu cells [30] and ovarian carcinoma cell growth [31], whereas high concentrations inhibit the cell proliferations, through assessing cell viability using a MTT assay. These data have stimulated interest into anti- or pro-cancer properties of ROS and RNS [22,32]. In clinical trials, inhibition of ROS generation, or delivery of high concentrations of NO via donor drugs or gene therapy has been attempted for cancer therapies [33,34] However, the roles of ROS and RNS on cell proliferation are debated [35-37].
Cell proliferation is usually evaluated, based on the metabolic activity of viable cells [38]. The MTT assay and the MTS assay are widely employed to assess cell proliferation in a range of situations, including investigations of the effects of ROS and RNS, and for evaluating new anti-cancer drug candidates [39,40]. The principle of the MTT assay is that tetrazolium salt (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) is converted into a formazan by mitochondrial enzymes [41,42]. It is assumed that mitochondria of viable cells produce these enzymes, but those of dead cells do not, and that the amount of formazan formed is directly proportional to the number of metabolically active cells in the culture [43,44].
One of the limitations of the MTT assay is that the formazin product produced is initially water insoluble. The sample needs to be dissolved in organic solvents, and then dissolved in water for a measurement with a spectrophotometer. The MTS assay is an improved version of the MTT assay, which formazan product is an aqueous soluble product that generated in the presence phenazine methosulfate (PMS) [45,46]. PMS is used as an electron coupling reagent [47]. The principle of the MTS assay is that 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2- (4-sulfophenyl)-2H-tetrazolium can be transformed to formazan by dehydrogenases in the inner mitochondrial membrane of cells [46].
The aim of this study was to investigate the potential effects of H2O2 and NO on HepG2 cell proliferation; and to determine whether synergistic effects occurred between NO and H2O2, and the effects of the anti-oxidants cPTIO and catalase on these. In this study, SNP and SNAP were used as NO donors; H2O2 as an oxidant; cPTIO as a NO scavenger; and catalase as an antioxidant enzyme. HepG2 cells were exposed to SNP and SNAP with concentrations ranging from 0 to 1000 μM, with or without 100 μM H2O2, 100 μM cPTIO and 100 U/mL catalase for 24 hours. Cell viabilities were assessed using the MTS assay. The hypothesis for these experiments was that in cultured HepG2 cells:
• Low concentrations of NO enhance proliferation, whereas high concentrations inhibit proliferation.
• Low concentrations of H2O2 enhance proliferation, whereas high concentrations inhibit proliferation.
• There are synergistic effects with the combination of SNP (or SNAP) and H2O2, cPTIO or catalase.
Materials and Methods
Materials
Sodium nitroprusside dehydrate (SNP, S0501), S-Nitroso-NAcetyl- Penicillamine (SNAP, N-3398), 2-(4-Carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide potassium salt (cPTIO, C221), catalase (C-3515) and trypan blue (T8154) were purchased from Sigma (St. Louis, MO, USA). Dulbecco’s Eagle minimum essential medium (DMEM) (11995-115), foetal bovine serum (0405), penicillin-streptomycin (0021) and trypsin (0187) were from Invitrogen (Mulgrave, Vic, Australia). Fungizone (list 43760) was from APOTHECON (Princeton, NJ, USA). Sodium hydrogen carbonate (NaHCO3, A475-500G) was from Analytical UNIVAR Reagent Ajax Finechem (Mt Wellington, New Zealand). Phosphate buffered saline (PBS) without Ca2+ and Mg2+ was from Queensland Institute of Medical Research (Brisbane, Australia).
HepG2 cells (a human hepatocellular carcinoma cell line, ATCC, HB8065) were obtained from Dr Grant Ramm’s laboratory at the Queensland Institute of Medical Research and originally sourced from the American Type Culture Collection (Manassas, VA, USA). CellTiter 96® AQueous Non-Radioactive Cell Proliferation Assay kit (G5421) was purchased in Promega Corporation (Madison, WI, USA).
Drug stock solutions
SNAP was dissolved in sterile DMSO, whereas SNP and cPTIO were dissolved in PBS to make stock solutions, then stored at - 20°C [48]. Solutions of H2O2 were freshly made at the time of use. 42 mg of MTS powder from the CellTiter 96® AQueous Non- Radioactive Cell Proliferation Assay kit was completely dissolved in 21 mL of Dulbecco’s phosphate buffered saline to prepare MTS solution. The MTS solutions were stored at - 20°C protected from light.
Cell culture
Cryovials containing HepG2 cells stored in a liquid nitrogen Dewar were removed to a water bath at 37°C to defrost the cells for 2 minutes. The cryovials were wiped with 70% ethanol and the contents of the tubes were poured into a 50 mL falcon tube containing warm complete medium. The falcon tube was spun at 500 × g for 5 minutes to form a small cream coloured pellet. The supernatant was poured off. Pellets were washed in fresh medium and resuspended in 10 mL of fresh complete medium. Pellets were separated by drawing through a sterile 21-gauge needle. Concentrations of the cells were determined using the trypan blue exclusion method, which cells were stained with trypan blue, and then, cells were counted in a hemocytometer under a microscope. Cell concentrations were adjusted to 1 × 106 cells/mL. 10 mL of this cell solution was transferred into a 75 cm2 tissue culture flask. The cells were cultured at 37°C in a humidified 5% CO2 and 95% air incubator. Cell cultures were checked microscopically daily to ensure that cells were healthy and growing as expected. Cultured medium was renewed every 3 days.
Cell subculture
Cells were subcultured after being grown to 70% confluence. Trypsin, versene, Dulbecco’s, PBS and complete DMEM medium were pre-warmed. The adhering cell monolayer was washed twice with a small volume of PBS at 37°C. Two ml of warm trypsin solution was added to the cultures covering the adhering cell layer. The flasks were placed in a 37°C warming tray for 2 minutes to dislodge cells. 30 mL of warm complete DMEM medium was added in the flask to neutralize the trypsin. Cells were then pelleted as described above and a similar process was undertaken to culture cells in 75 cm2 tissue culture flasks.
Cell seeding
When cells grew to 60% confluence in 75 cm2 flasks, they were dislodged with trypsin as described above, washed and resuspended to make 50 mL of cell solution with a concentration of 2 × 105 cells/mL. 100 μL of the cell solution was added into each well of 96-well plates. These cells were cultured for 48 hours at 37°C.
Treatment
Prior to use, working solutions of H2O2, cPTIO, SNP and SNAP were prepared in PBS to concentrations ranging from 10 μM to 10 mM. 10 μL of the appropriate working solution was added into each culture well containing 90 μL of cell solution by 10- fold serial dilution. In the first set of experiment, cells were exposed to concentrations of 0, 1, 10, 50, 100, 200 and 1000 μM SNP, SNAP, H2O2 or cPTIO. In a second set of experiments, cells were exposed to 0, 1, 10, 50, 100, 200 and 1000 μM SNP or SNAP with or without 100 μM of H2O2, 100 μM of cPTIO or 100 U/mL of catalase for 24 hours at 37°C, 5% CO2, in a humidified incubator.
Cell proliferation assay
Immediately before cell viability was measured, 2 mL of MTS solution and 100 μL of PMS solution were removed from the freezer, thawed in a 37°C water bath and mixed evenly using aseptic techniques. Once cells were treated with the chemicals for 24 hours, 20 μL of the combined MTS/PMS solution were added into each well of the 96-well plate containing the cell suspension and then incubated for 2 hours at 37°C, 5% CO2 in a humidified incubator. The absorbance was recorded at 490 nm using an ELISA plate reader. The viability of cells was calculated for each assay using the formula:
Percentages of cell proliferation=100 × Absorbance values of test sample/Absorbance values of control sample Equation 1
Statistical analysis
All statistical analyses and graphs were performed using GraphPad Prism 6.0 from GraphPad Software, Inc (Avenida de la Playa La Jolla, USA). Statistical differences of proliferations rate of HepG2 cells treated with SNP, SNAP, H2O2 or cPTIO compared to those of HepG2 cells were not treated with these chemicals were determined using one-way analysis of variance (ANOVA) followed by Dunnett's Multiple Comparison Test. Significant differences of proliferation rates of various concentration points between SNP (or SNAP) with100 μM H2O2, 100 μM cPTIO or 100 U/mL catalase and SNP (or SNAP) alone were determined with Bonferroni and Šídák methods. P values<0.05 were considered significantly different. Each experimental condition was performed in triplicate. All experiments were repeated for three times. Data were expressed as mean % ± standard deviation (SD).
Results
Proliferation of HepG2 cells treated with SNP, SNAP, H2O2 or cPTIO
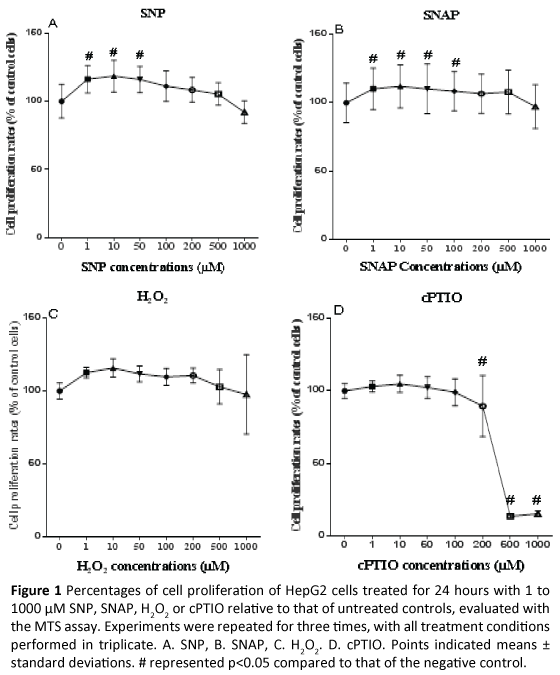
HepG2 cells were treated with 0 to 1000 μM SNP, SNAP, H2O2 or cPTIO for 24 hours. Cell proliferations were assessed with the MTS assay. Statistical differences of proliferation rates of HepG2 cells treated with various concentrations of SNP, SNAP, H2O2 or cPTIO compared to those of negative controls that were not treated with these chemicals were determined using one-way analysis of variance followed by a Dunnett's Multiple Comparison Test. A p value of less than 0.05 was considered statistically significant. Cell proliferation of HepG2 cells significantly increased from 1 to 50 μM SNP, but there was no significant difference from 100 to 1000 μM SNP compared to negative controls (Figure 1A). There was a significant increase in cell proliferation at doses of SNAP between 1 to 100 μM SN AP, but there was no significant difference from 200 to 1000 μM SNAP compared to negative controls (Figure 1B). There was a trend that low concentrations (1 to 200 μM) of H2O2 enhanced cell proliferation, but there was no a statistically significant change from 500 to 1000 μM H2O2 compared to negative controls (Figure 1C). High concentrations (200 to 1000 μM) of cPTIO reduced absorbance values, in particular, 500 and 1000 μM cPTIO reduced 80% absorbance values (Figure 1D).
Figure 1: Percentages of cell proliferation of HepG2 cells treated for 24 hours with 1 to 1000 µM SNP, SNAP, H2O2 or cPTIO relative to that of untreated controls, evaluated with the MTS assay.Experiments were repeated for three times, with all treatment conditions performed in triplicate. A. SNP, B. SNAP, C. H2O2. D.cPTIO. Points indicated means ± standard deviations. # represented p<0.05 compared to that of the negative control.
HepG2 cell proliferation induced by SNP without or with H2O2, cPTIO, or catalase
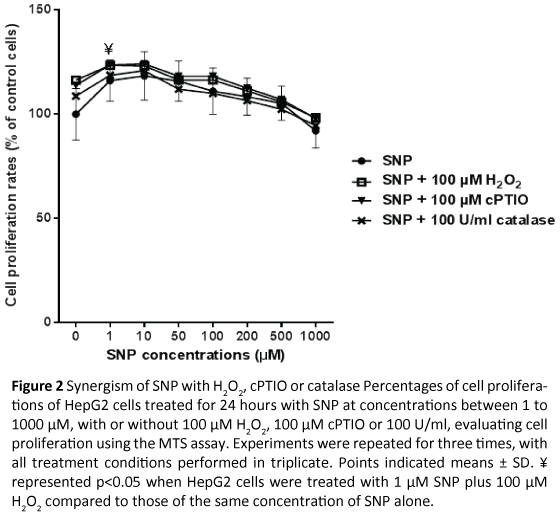
HepG2 cells were treated with 0 to 1000 μM SNP with or without 100 μM H2O2, 100 μM cPTIO or 100 U/mL catalase for 24 hours, before cell proliferation was evaluated with the MTS assay. A similar trend to SNP alone was observed when SNP at various concentrations was combined with 100 μM H2O2, 100 μM cPTIO or 100 U/mL catalase, which low concentrations of SNP enhanced cell proliferations (Figure 2). A Bonferroni and Šídák’s Multiple Comparison Test indicated that there was a statistically significantly differences between proliferations of HepG2 cells treated with 1 μM SNP alone or with 100 μM H2O2. This was not seen for other concentrations of SNP ± 100 μM H2O2. Moreover, at various concentration points of SNP, there was no significant difference between proliferation rates of HepG2 cells treated with SNP alone or when combined with 100 μM cPTIO or 100 U/ mL catalase.
Figure 2: Synergism of SNP with H2O2, cPTIO or catalasePercentages of cell proliferations of HepG2 cells treated for 24 hours with SNP at concentrations between 1 to 1000 µM, with or without 100 µM H2O2, 100 µM cPTIO or 100 U/ml, evaluating cell proliferation using the MTS assay. Experiments were repeated for three times, with all treatment conditions performed in triplicate. Points indicated means ± SD. ¥ represented p<0.05 when HepG2 cells were treated with 1µM SNP plus 100 µM H2O2 compared to those of the same concentration of SNP alone.
HepG2 cell proliferation mediated by SNAP with or without H2O2, cPTIO or catalase
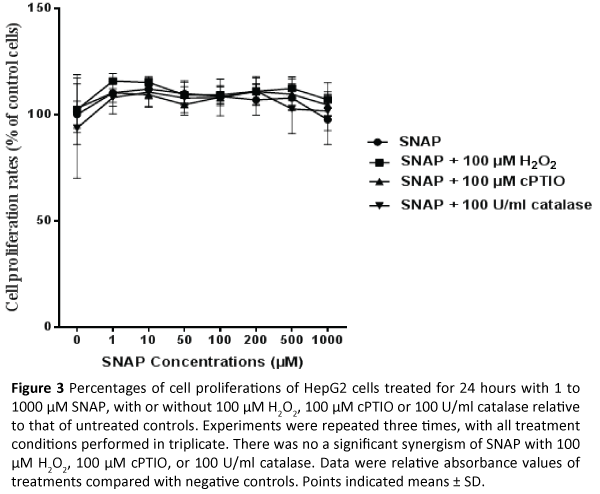
HepG2 cells were treated with SNAP with or without 100 μM H2O2, 100 μM cPTIO or 100 U/mL catalase for 24 hours, with cell proliferation measured by the MTS assay. A Bonferroni and Šídák’s Multiple Comparison Test indicated that there were not statistically significant differences between proliferations of HepG2 cells treated with various concentrations of SNAP alone and those of HepG2 cells treated with SNAP plus 100 μM H2O2, 100 μM cPTIO or 100 U/mL catalase (Figure 3).
Figure 3: Percentages of cell proliferations of HepG2 cells treated for 24 hours with 1 to 1000 µM SNAP, with or without 100 µM H2O2, 100 µM cPTIO or 100 U/ml catalase relative to that of untreated controls. Experiments were repeated three times, with all treatment conditions performed in triplicate. There was no a significant synergism of SNAP with 100 µM H2O2, 100 µM cPTIO, or 100 U/ml catalase. Data were relative absorbance values of treatments compared with negative controls. Points indicated means ± SD.
Discussion
This study did not support the hypothesis that low concentrations of NO enhanced proliferation, and that high concentrations inhibited proliferation. The MTS assay revealed that (1, 10, 50 μM) SNP and (1, 10, 50, 100 μM) SNAP significantly enhanced HepG2 cell proliferation, but (100 to 1000 μM) SNP and (200 to 1000 μM) SNAP showed a non-significant trend to inhibit cell proliferation (Figure 1A and 1B). These dada indicated that SNAP, in higher dose (100 μM), significantly enhanced cell proliferations, but 100 μM SNP did not. However, my research revealed that 100 μM SNAP released 37 ± 14 μM NO whereas 100 μM SNP released 5.8 ± 1.8 μM NO. SNAP released more NO than SNP, and both of them released NO in dose-dependent manner. A previous study also revealed that SNAP released more NO than SNP [49]. It is to say that the cell proliferations indicated by the MTS assay in this study were not relative to NO concentrations.
This study did not support the hypothesis that low concentrations of H2O2 enhance proliferation, whereas high concentrations inhibit proliferation. The MTS assay revealed a trend that low concentrations of H2O2 enhanced HepG2 cell proliferation, and high concentrations of H2O2 inhibit cell proliferation, but there were no significant differences (Figure 1C). The result was different from that of a previous study that for keratinocytes exposed to up to 1000 μM H2O2 for 24 hours, where the relative absorbance increase to 102 to 115% of control for 100 to 500 μM H2O2, but reduced by 60% at 1000 μM H2O2 [25].
This study first proposed that cPTIO interfered with the MTS assay. The MTS assay revealed that 500 and 1000 μM cPTIO reduced absorbance values to 15.2 and 13.4% of that of negative controls respectively (Figure 1D). Thus, it appears that in excess of 80% of HepG2 cells treated with 500 or 1000 μM cPTIO died. However, my pervious experiments demonstrated that the modified trypan blue method and the LDH assays did not identify major toxic effect of 500 and 1000 μM cPTIO on HepG2 cells. This suggests that the low MTS assay read out with concentrations of cPTIO was due to an effect of cPTIO on the MTS assay rather than HepG2 cell death. Previous studies have shown that cPTIO is an oxidant [48,50] and has a similar structure to MTS and MTT [46,51]. Thus, at high concentrations cPTIO competes with MTS and MTT for electrons, leading to reduced transformations of MTS and MTT to formazan, leading to decrease of absorbance values. There are similar observations in previous studies. For example, 500 μM cPTIO significantly decreased absorbance values of human breast carcinoma MDA-MB-468 cells after a 48 hour treatment, determined using the MTT assay [52]. According to above discussion, these data cannot be inferred that high concentrations of cPTIO induce large cell death.
This study did not support the hypothesis, that there was adequate significant evidence of a synergistic interaction between NO and H2O2, cPTIO or catalase on HepG2 cell proliferation. Figure 2 did not reveal significant difference between SNP and SNP plus 100 μM H2O2, 100 μM cPTIO, or 100 U/ml catalase (except 1 μM SNP plus 100 μM H2O2). Figure 3 did not reveal significant difference between SNAP and SNAP plus 100 μM H2O2, 100 μM cPTIO, or 100 U/mL catalase either. At 1μM SNP, there was significant difference between SNP and SNP plus 100 μM H2O2, but at a large amount of other concentrations of SNP (or SNAP), there was no significant difference between SNP (or SNAP) and SNP (or SNAP) plus100 μM H2O2.
This study has several limitations. Theoretically, the tetrazolium salts of the MTT and MTS assay are reduced to colorimetric formazan by succinate dehydrogenase, with absorbance being proportional to the number of viable cells. However, accumulated data have shown that a number of parameters interfere with the MTT and MTS assays.
1. The conversion of tetrazolium to formazan is oxidoreduction, but release of NO from SNP or SNAP is also oxidoreduction. Therefore, transfer of electrons can occur between these agents, potentially interfering with the assay. Moreover, the mechanisms of the bio-reduction of tetrazolium [53] and the release of NO by SNP [54,55] or SNAP [56,57] are not completely clear.
2. SNP, SNAP and H2O2 decay at different rates. It has been reported that 100 μM H2O2 decays rapidly to zero within 1 hour in the presence of cells in the media, while NO released by SNAP increases in a time-dependant manner [58]. This means that H2O2 may not react with NO before t completely decays.
3. Some chemical agents may interact with the MTT and MTS assays without affecting actual cell viability. Recent reports have demonstrated that cholesterol [59], plant extracts [60], emodin [61], ascorbic acid [62] and flavonoids [63] chloroquine [38] all may interfere with the MTT assay.
4. In addition to mitochondrial dehydrogenases and oxidoreductases, enzymes in the cytoplasm and in nonmitochondrial membranes can also reduce tetrazolium to formazan [43,64]. Many other reductants, e.g., NADH, NADPH, succinate and pyruvate, are also capable of reducing tetrazolium in the presence [53] or absence of enzymes [65,66].
Summary
The MTS assay in this assay revealed that low concentrations of SNP and SNAP can significantly enhance HepG2 cell proliferation, whereas the changes with high concentrations were not statistically significantly different from control cells. H2O2 exerted a trend that low concentrations enhanced the cell growth whereas high concentrations inhibited cell growth, but there was no statistically significant difference. 100 μM H2O2, 100 μM cPTIO or 100 U/mL catalase did not have evident synergisms with NO. However, these data should be carefully explained due to interferences of a number of parameters with the MTS assays. Furthermore, due to the interference, conclusions in earlier studies on pro-cancer or anti-cancer roles of reactive oxygen and nitrogen species and their scavengers are uncertain.
Acknowledgement
This study is one part of my PhD thesis. I would like to thank my Principle Advisor Associate Professor Graeme Macdonald for his time, patience, expertise, guidance and mentorship throughout my PhD studies, enabling me to complete my thesis. I am also genuinely grateful for the assistance that my Associate Advisor, Professor Grant Ramm lets me be a member of his laboratory. I would like to thank Dr Leon Hugo from the Mosquito Control Group of the Queensland Institute of Medical Research for correcting my thesis manuscript and giving a great deal of invaluable advice. It is my pleasure to thank lab members of the Hepatic Fibrosis Group, Iron Metabolism Group and Membrane Transport Group at the Queensland Institute of Medical Research for their friendship, support, guidance, encouragement, anecdotes, advice, and amusement during my candidature. I would like to thank members of the Herston Library of the University of Queensland for their help. I would like to acknowledge the University of Queensland for providing me with financial support for my studies in the forms of an Australian Postgraduate Award scholarship.
References
- Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144: 646-674.
- Balmer ML, JF Dufour, KM McMasters (2011) Biology of Hepatocellular Carcinoma Hepatocellular Carcinoma. Springer 21-34.
- Tsai WL, Chung RT (2010) Viralhepatocarcinogenesis. Oncogene 29: 2309-2324.
- Muriel P (2009) Role of free radicals in liver diseases. HepatolInt 3: 526-536.
- Pervin S, Rajan S, Suvajit S, Gautam C (2010) Dual Role of Nitric Oxide in Cancer Biology, in Nitric Oxide (NO) and Cancer 39-57.
- Finkel T (2011) Signal transduction by reactive oxygen species. J Cell Biol 194: 7-15.
- Monga SPS, Cubero FJ, TrautweinC (2011) Oxidative Stress and Liver Injury, in Molecular Pathology of Liver Diseases. Springer,pp: 427-435.
- Federico A, Morgillo F, Tuccillo C, Ciardiello F, Loguercio C (2007) Chronic inflammation and oxidative stress in human carcinogenesis. Int J Cancer 121: 2381-2386.
- Pârvu AE, Negrean V, Pleaca-Manea L, Cosma A, Draƒghici A, et al. (2005) Nitric oxide in patients with chronic liver diseases. Rom J Gastroenterol 14: 225-230.
- Rosa MP, Frau M,Francesco F (2010) Prognostic Significance of iNOS in Hepatocellular Carcinoma. Nitric Oxide (NO). Cancer,pp: 309-328.
- Ziech D, Franco R, Pappa A, Panayiotidis MI (2011) Reactive Oxygen Species (ROS)--Induced genetic and epigenetic alterations in human carcinogenesis. Mutat Res 711: 167-173.
- Sedelnikova OA(2010) Role of oxidatively induced DNA lesions in human pathogenesis. Mutation Research/Reviews in Mutation Research 704: 152-159.
- Fukumura D, Kashiwagi S, Jain RK (2006) The role of nitric oxide in tumour progression. Nat Rev Cancer 6: 521-534.
- Jain S, Singhal S, Lee P, Xu R (2010) Molecular genetics of hepatocellular neoplasia. Am J Transl Res 2: 105-118.
- Laurent-Puig P, Zucman-Rossi J (2006) Genetics of hepatocellular tumors. Oncogene 25: 3778-3786.
- Veal EA, Day AM, Morgan BA (2007) Hydrogen peroxide sensing and signaling. Mol Cell 26: 1-14.
- Gough DR, Cotter TG (2011) Hydrogen peroxide: a Jekyll and Hyde signalling molecule. Cell Death Dis 2: e213.
- Miki H, Funato Y (2012) Regulation of intracellular signalling through cysteine oxidation by reactive oxygen species. Biochem 151: 255-261.
- Blaise GA, Gauvin D, Gangal M, Authier S (2005) Nitric oxide, cell signaling and cell death. Toxicology 208: 177-192.
- Jia M, Mateoiu C, Souchelnytskyi S (2011) Protein tyrosine nitration in the cell cycle. BiochemBiophys Res Commun 413: 270-276.
- Liaudet L, Vassalli G, Pacher P (2009) Role of peroxynitrite in the redox regulation of cell signal transduction pathways. Front Biosci (Landmark Ed) 14: 4809-4814.
- Liou GY, Storz P (2010) Reactive oxygen species in cancer. Free Radic Res 44: 479-496.
- Dickinson BC, Chang CJ (2011) Chemistry and biology of reactive oxygen species in signaling or stress responses. Nat ChemBiol 7: 504-511.
- Ridnour LA, Thomas DD, Switzer C, Flores-Santana W, Isenberg JS, et al. (2008) Molecular mechanisms for discrete nitric oxide levels in cancer. Nitric Oxide 19: 73-76.
- Loo AEK, Ho R, Halliwell B (2011) Mechanism of hydrogen peroxide-induced keratinocyte migration in a scratch-wound model. Free Radical Biology and Medicine 51: 884-892.
- Burdon RH, Gill V (1993) Cellularly generated active oxygen species and HeLa cell proliferation. Free Radic Res Commun 19: 203-213.
- Ohguro N, Fukuda M, Sasabe T, TanoY (1999) Concentration dependent effects of hydrogen peroxide on lens epithelial cells. British Journal of Ophthalmology 83: 1064-1068.
- Polytarchou C, Hatziapostolou M, Papadimitriou E (2005) Hydrogen Peroxide Stimulates Proliferation and Migration of Human Prostate Cancer Cells through Activation of Activator Protein-1 and Up-regulation of the Heparin Affin Regulatory Peptide Gene. Journal of Biological Chemistry 280: 40428-40435.
- Rabkin SW, Kong JY (2000) Nitroprusside induces cardiomyocyte death: interaction with hydrogen peroxide.Am J Physiol Heart CircPhysiol 279: H3089-3100.
- Fetz V, Bier C, Habtemichael N, Schuon R, Schweitzer A, et al. (2009) Inducible NO synthase confers chemoresistance in head and neck cancer by modulating survivin. Int J Cancer 124: 2033-2041.
- Engels K,Knauer SK, Loibl S, Fetz V, Harter P, et al.(2008) NO signaling confers cytoprotectivity through the survivin network in ovarian carcinomas. Cancer Research68: 5159-5166.
- Robert C, Ridnour LA, Glynn SA, Switzer CH, Flores-Santana W, et al. (2010) Nitric Oxide and Cancer: An Overview. Nitric Oxide (NO) and Cancer, pp: 3-20.
- Hiroyasu Y, Kazuhiro Y, Katsutoshi N, Tadashi M, Takahiko S, et al. (2010) Therapeutic Applications of Nitric Oxide for Malignant Tumor in Animal Models and Human Studies. Nitric Oxide (NO) and Cancer,pp: 419-441.
- Coulter JA, McCarthy HO, Xiang J, Roedl W, Wagner E, et al. (2008) Nitric oxide--a novel therapeutic for cancer. Nitric Oxide 19: 192-198.
- Chang MC, Chan CP, Wang YJ, Lee PH, Chen LI, et al. (2007) Induction of necrosis and apoptosis to KB cancer cells by sanguinarine is associated with reactive oxygen species production and mitochondrial membrane depolarization. Toxicology and Applied Pharmacology 218:143-151.
- Halliwell B (2007) Oxidative stress and cancer: have we moved forward? Biochem J 401: 1-11.
- Masini E, Fabio C, Rosanna M, Salvatore C (2010) Nitric Oxide Expression in Cancer. Nitric Oxide (NO) and Cancer,pp: 59-82.
- Weyermann J, Lochmann D, Zimmer A (2005) A practical note on the use of cytotoxicity assays. Int J Pharm 288: 369-376.
- Hayon T, Dvilansky A, Shpilberg O, Nathan I (2003) Appraisal of the MTT-based assay as a useful tool for predicting drug chemosensitivity in leukemia. Leuk Lymphoma 44: 1957-1962.
- Nomura N, Kaoru S, Mayako K, Pi-Chao W, Tadao O, et al. (1996) Improved MTT(3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) assay for the measurement of viable animal cell number in porous cellulose carriers. Biotechnology Techniques 10:883-888.
- Slater TF, Sawyer B, Sträuli U(1963) Studies on succinate-tetrazoliumreductase systems: III. Points of coupling of four different tetrazolium salts III. Points of coupling of four different tetrazolium salts. BiochimBiophysActa. 77: 383-393.
- Sylvester PW (2011) Optimization of the tetrazolium dye (MTT) colorimetric assay for cellular growth and viability. Methods MolBiol 716: 157-168.
- Liu Y, Peterson DA, Kimura H, Schubert D (1997) Mechanism of cellular 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reduction. J Neurochem 69: 581-593.
- Butler M, Spearman M, PörtnerR (2007) Cell Counting and Viability Measurements.Animal Cell Biotechnology24: 205-222.
- O'Toole S, Sheppard BL, McGuinness EP, Gleeson NC, Yoneda M,et al. (2003) The MTS assay as an indicator of chemosensitivity/resistance in malignant gynaecologicaltumours. Cancer Detection and Prevention 27: 47-54.
- Barltrop JA, Terence CO, Ann HC, Joseph GC (1991)5-(3-carboxymethoxyphenyl)-2-(4,5-dimethylthiazolyl)-3-(4-sulfophenyl)tetrazolium, inner salt (MTS) and related analogs of 3-(4,5-dimethylthiazolyl)-2,5-diphenyltetrazolium bromide (MTT) reducing to purple water-soluble formazans As cell-viability indicators. Bioorganic& Medicinal Chemistry Letters 1: 611-614.
- Leverone MR, Owenb TC, Tiedera FS, Stewartc GJ, Lim DV (1996) Resting-cell dehydrogenase assay measuring a novel water-soluble formazan detects catabolic differences among cells. Journal of Microbiological Methods 25:49-55.
- Amano F,NodaT (1995) Improved detection of nitric oxide radical (NO-) production in an activated macrophage culture with a radical scavenger, carboxy PTIO, and Griess reagent. FEBS Letters 368: 425-428.
- Motterlini R, Motterlini R, Foresti R, Intaglietta M, Winslow RM (1996) NO-mediated activation of hemeoxygenase: endogenous cytoprotection against oxidative stress to endothelium. Am J Physiol Heart CircPhysiol 270: H107-114.
- Akaike, T, Masaki Y, Yoichi M, Keizo S, Masahiro K,et al. (1993) Antagonistic action of imidazolineoxyl N-oxides against endothelium-derived relaxing factor/.bul.NO (nitric oxide) through a radical reaction. Biochemistry 32: 827-832.
- Lakshmi V,ZenserT (2007) 2-(4-Carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide potentiates nitrosation of a heterocyclic amine carcinogen by nitric oxide. Life Sciences 80: 644-649.
- Lo HW, Hsu SC, Ali-Seyed M, Gunduz M, Xia W, et al. (2005) Nuclear interaction of EGFR and STAT3 in the activation of the iNOS/NO pathway. Cancer Cell 7: 575-589.
- Berridge MV, Herst PM, Tan AS (2005) Tetrazolium dyes as tools in cell biology: new insights into their cellular reduction. BiotechnolAnnu Rev 11: 127-152.
- Grossi L,D'AngeloS (2005) Sodium Nitroprusside: Mechanism of NO Release Mediated by Sulfhydryl-Containing Molecules. Journal of Medicinal Chemistry 48: 2622-2626.
- Williams DL (2003) A chemist's view of the nitric oxide story. Org BiomolChem 1: 441-449.
- Bainbrigge N, Butler AR, GorbitzCH (1997)The thermal stability of S-nitrosothiols: Experimental studies and ab initio calculations on model compounds. Journal of the Chemical Society-Perkin Transactions 2: 351-353.
- Anthony AR, RhodesP (1997) Chemistry, Analysis, and Biological Roles ofS-Nitrosothiols. Analytical Biochemistry 249: 1-9.
- Wang JY, Shum AYC, HoYJ (2003) Oxidative neurotoxicity in rat cerebral cortex neurons: Synergistic effects of H2O2 and NO on apoptosis involving activation of p38 mitogen-activated protein kinase and caspase-3. Journal of Neuroscience Research 72: 508-519.
- Ahmad S (2006) Cholesterol Interferes with the MTT Assay in Human Epithelial-Like (A549) and Endothelial (HLMVE and HCAE) Cells. International Journal of Toxicology 25: 17-23.
- Bruggisser R(2002) Interference of Plant Extracts, Phytoestrogens and Antioxidants with the MTT Tetrazolium Assay. Planta Med 68: 445-448.
- Wang X, Ge J, Wang K, Qian J, Zou Y (2006) Evaluation of MTT assay for measurement of emodin-induced cytotoxicity. Assay Drug DevTechnol 4: 203-207.
- Chakrabarti R, Kundu S, Kumar S, Chakrabarti R (2000) Vitamin A as an enzyme that catalyzes the reduction of MTT to formazan by vitamin C. J Cell Biochem 80: 133-138.
- Peng L, Wang B, Ren P (2005) Reduction of MTT by flavonoids in the absence of cells. Colloids Surf B Biointerfaces 45: 108-111.
- Bernas T, Dobrucki J (2002) Mitochondrial and nonmitochondrial reduction of MTT: interaction of MTT with TMRE, JC-1, and NAO mitochondrial fluorescent probes. Cytometry 47: 236-242.
- Picker SD,FridovichI (1984)On the mechanism of production of superoxide radical by reaction mixtures containing NADH, phenazinemethosulfate and nitrobluetetrazolium. Archives of Biochemistry and Biophysics 228: 155-158.
- Van N, ButcherRG (1989)The involvement of superoxide anions in the nitro blue tetrazolium chloride reduction mediated by NADH and phenazinemethosulfate. Analytical Biochemistry176: 170-174.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences